About
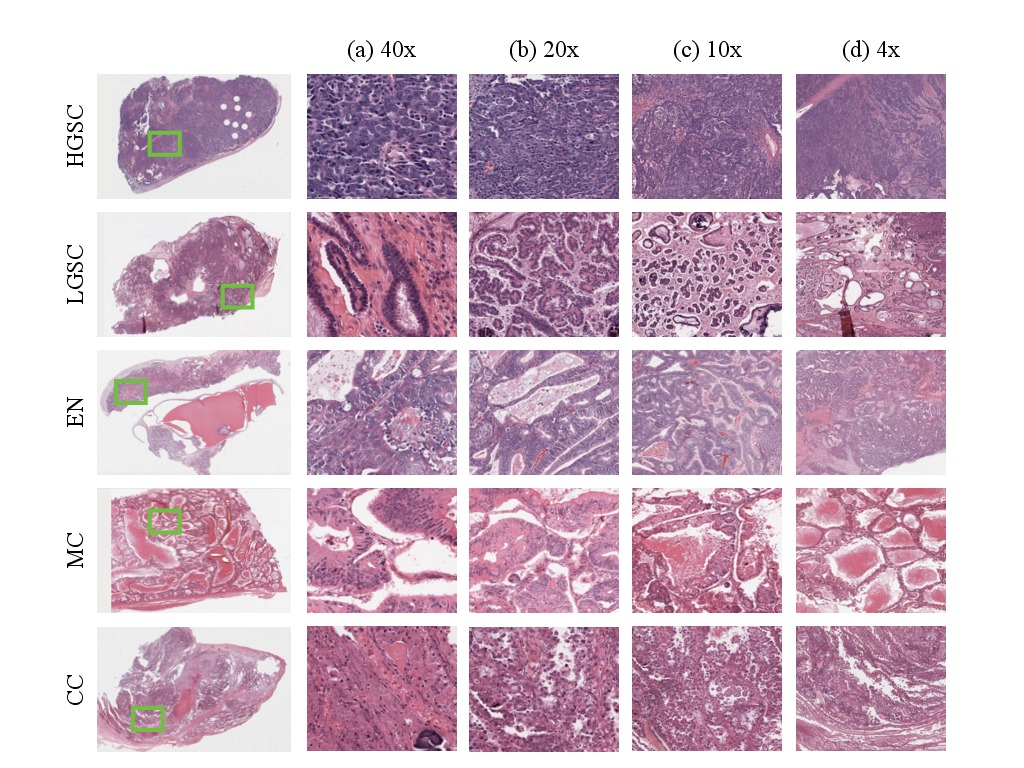
The ovarian carcinomas (OC) dataset is a growing collection of whole slide histopathology images digitzed from biopsy sections of five ovarian carcinoma subtypes: high grade serous (HGSC), low grade serous (LGSC), endometrioid (EN), mucinous (MC) and clear cell carcinomas (CC). At present, the collection includes slides from 80 different patients equally split in training and test sets.
This collection of whole slide images was acquired in the context of a Transcanadian study on the reproducibility of ovarian carcinomas subtyping accross 6 different pathology centers. Each whole slide image was digitized with an AperioScope scanner at 40x magnification and was selected by expert pathologists to cover as much of the lesion as possible from a selection of tissue slides. In addition each image has associated meta-data (immunocytology results) provided along with the final diagnosis.
The dataset was introduced to evaluate clinicians' agreement and diagnostic reproducibility then extended to evaluate automatic multiclass classification systems for ovarian carcinomas, where the goal is to automatically predict a carcinoma subtype for each whole slide image.
Benchmarks
The following papers proposed automatic systems for ovarian carcinoma subtypes classification using this dataset:
Clinically-Inspired Automatic Diagnosis of Ovarian Carcinoma Subtypes
A. BenTaieb, M. Nosrati, H. Li-Chang, D. Huntsman and G. Hamarneh.
Journal of Pathology Informatics 2016
[pdf] [code]
Automatic Diagnosis of Ovarian Carcinomas via Sparse Multiresolution Tissue Representation
A. BenTaieb, H. Li-Chang, D. Huntsman and G. Hamarneh.
MICCAI 2015
[pdf]
A Structured Latent Model for Ovarian Carcinoma Subtyping from Histopathology Slides
A. BenTaieb, H. Li-Chang, D. Huntsman and G. Hamarneh.
Medical Image Analysis 2017
[pdf]
[code]
Acknowledgements
This dataset is for academic, non-commercial use only.
If you use this dataset in a publication, please cite the following paper:
@article{kobel2010diagnosis,
title={Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study},
author={K{\"o}bel, Martin and Kalloger, Steve E and Baker, Patricia M and Ewanowich, Carol A and Arseneau, Jocelyne and Zherebitskiy,
Viktor and Abdulkarim, Soran and Leung, Samuel and Duggan, M{\'a}ire A and Fontaine, Dan and others},
journal={The American journal of surgical pathology},
volume={34},
number={7},
pages={984--993},
year={2010},
publisher={LWW}
}
Download
Please provide your email address and affiliation in the form below to receive the password needed to access the dataset and to be notified of any major changes to the dataset (updates, bug fixes, etc.).
After filling the form above, proceed with the following steps to access the dataset:
1) Download all files linked below
2) Use the following command to merge all files into a single folder
(WARNING: total uncompressed file size ~67 GB) containing all histopathology slides in svs format: cat data_part* > total_data.zip
3) Use the password sent to your inbox to uncompress the final archive file. (no longer needed)

